Note. This is updated from a previous blog post I published a long time ago in robotic healthcare on January 28, 2019.
According to a 2020 US census. 62 million Americans (19% of the population) identified as Hispanics where cancer continues to be their leading cause of death, accounting for 20% mortality.
Interestingly, although Hispanics are less likely to suffer from the most common cancers (e.g., lung, breast) than non-Hispanic Whites, they have a higher risk for those less common cancers that are also associated with infectious agents (e.g., liver, cervix).
Histology, or also known as microscopic anatomy, is a branch of biology necessary for visualizing patient samples taken from a tissue or skin lesion infected by a pathogen (microorganism that causes disease) or from a cancer tumor.
It remains as the core technique of classifying many rare tumors like glioblastoma (a type of brain cancer) because unlike common tumors, there is a lack of molecular identifiers that would allow technological developments to assess them without needing visual appraisal of cellular alterations.
The problem with histology
As it depends on visual observations, these can be vary from person to person leading to different classifications based on different assessments, thus introducing bias.
Along with this human variation, there is also the challenge that although tumors can have similar histology, they can still progress in different ways, and so the other way around, where tumors with different microscopic characteristics can progress the same way.
In previous research studies (1, 2) for example, this inter-observer variability in histopathological diagnosis has been reported in Central Nervous System (CNS) tumors like diffuse gliomas (brain tumors initiating in a type of brain cells called glial cells), ependymomas ( brain tumors initiating in the ependymoma), and supratentorial primitive neuroectodermal tumors ( occurring mostly in children starting in the cerebrum).
To try to address this problem, some molecular groupings have been updated into the World Health Organization (WHO) classification, but at the time of this writing only for selected tumors such as medulloblastoma.
This diagnostic variation and uncertainty provide a challenge to decision-making in clinical practice that can have a major effect on the survival of a cancer patient.
Therefore, Capper and colleagues decided to train their machine learning algorithm focusing not on complex visual assessments, but on the most studied epigenetic event in cancer, DNA methylation.
Histology vs DNA methylation
Within our hard-working cells, epigenetic (epi meaning above, genetic meaning genes) modifications do not affect the DNA sequence or genetic coding that contain the instructions our cells need to work.
However, this type of modification does alter the expression of the resulting genes produced from this genetic coding and end up affecting how are our cells do the work. In DNA methylation, a chemical group called a methyl group is bound to the DNA, and this feature is diverse in specific cancers which allow for innovative diagnostics to classify them.
Compared with histology, epigenome analysis of DNA methylation in cancer allows for an unbiased diagnostic approach, and thus David Capper and colleagues fed their innovative cancer diagnostic computer genome-wide methylation data from samples of almost all CNS tumors typed under WHO classification and their work was published in the scientific journal Nature in 2018 .
Machine Learning + DNA methylation
Capper et al. (2018) used the machine learning algorithm Random Forest (RF), as it combines several weak classifiers to improve the accuracy of the prediction, and trained it to recognize methylation patterns in the provided already histological-classified samples via supervised machine learning and find naturally occurring tumor patterns by itself to assign the samples based on this pattern category.
Capper and his colleagues then used the computer to classify 1,104 test cases which has been diagnosed by pathologists using standard histological and molecular way. An overview of their findings showcases their interesting results:
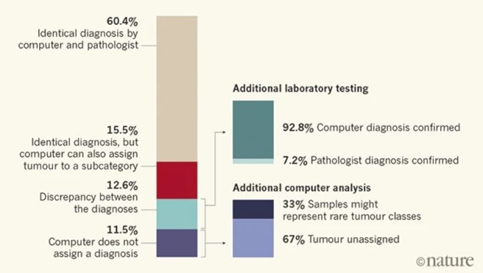
In 12.6% of the cases, the computer and pathologist diagnosis did not match, but after further laboratory testing involving a technique called gene sequencing that allows to see DNA changes at the genetic level, 92.8% of these unmatched tumors were found to correctly match the computers and not the pathologist’s assessment. Furthermore, 71% of these were computationally assigned a different tumor grade, which affect treatment delivery.
The Future
Despite this machine learning innovation, today histology remains as the indispensable method for accessible and universal tumor classification. However, the approach developed by Capper et al. (2018) complements and, in some cases such as rare tumor classification, outrivals histological microscopic examination. As this platform further develops in present laboratories, the future of cancer classification might prove one of utmost accuracy and unbiased approach by the combination of visual inspection and molecular analysis.

